Examples¶
Subsetting and selecting data¶
Let’s open a WRF model output file:
In [1]: import salem
In [2]: from salem.utils import get_demo_file
In [3]: ds = salem.open_xr_dataset(get_demo_file('wrfout_d01.nc'))
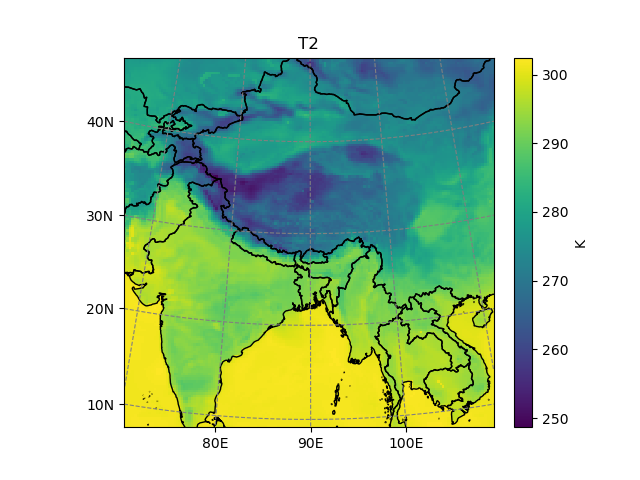
Let’s take a time slice of the variable T2 for a start:
In [4]: t2 = ds.T2.isel(Time=2)
In [5]: t2.salem.quick_map()
Out[5]: <salem.graphics.Map at 0x7f97fea59360>
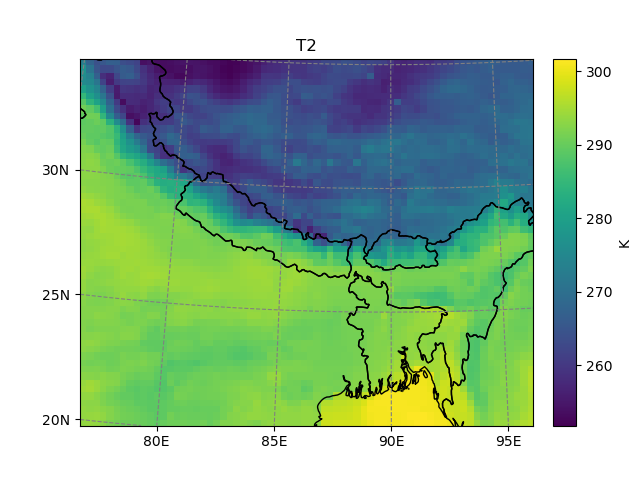
Although we are on a Lambert Conformal projection, it’s possible to subset the file using longitudes and latitudes:
In [6]: t2_sub = t2.salem.subset(corners=((77., 20.), (97., 35.)), crs=salem.wgs84)
In [7]: t2_sub.salem.quick_map()
Out[7]: <salem.graphics.Map at 0x7f980d2c3a00>
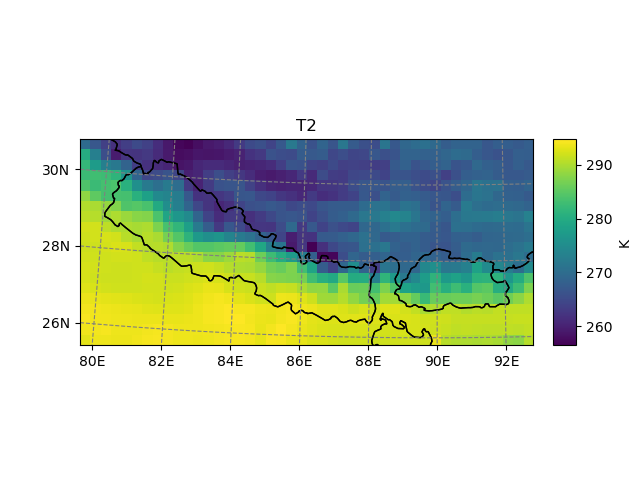
It’s also possible to use geometries or shapefiles to subset your data:
In [8]: shdf = salem.read_shapefile(get_demo_file('world_borders.shp'))
In [9]: shdf = shdf.loc[shdf['CNTRY_NAME'].isin(['Nepal', 'Bhutan'])] # GeoPandas' GeoDataFrame
In [10]: t2_sub = t2_sub.salem.subset(shape=shdf, margin=2) # add 2 grid points
In [11]: t2_sub.salem.quick_map()
Out[11]: <salem.graphics.Map at 0x7f97fedc7520>
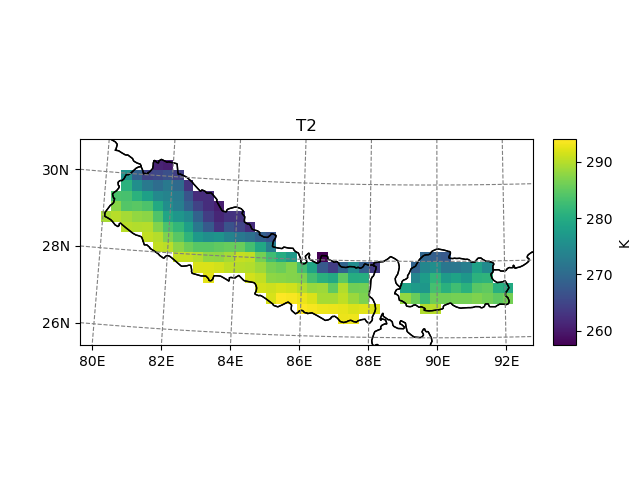
Based on the same principle, one can mask out the useless grid points:
In [12]: t2_roi = t2_sub.salem.roi(shape=shdf)
In [13]: t2_roi.salem.quick_map()
Out[13]: <salem.graphics.Map at 0x7f980d2c0790>
Plotting¶
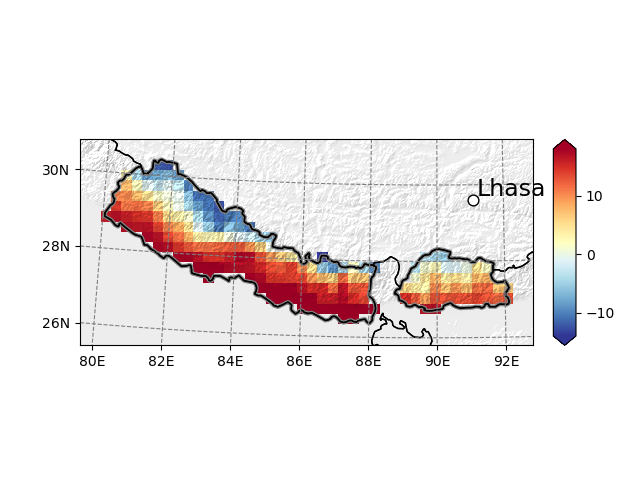
Maps can be embellished with topographical shading, points of interest, and more:
In [14]: smap = t2_roi.salem.get_map(data=t2_roi-273.15, cmap='RdYlBu_r', vmin=-14, vmax=18)
In [15]: _ = smap.set_topography(get_demo_file('himalaya.tif'))
In [16]: smap.set_shapefile(shape=shdf, color='grey', linewidth=3)
In [17]: smap.set_points(91.1, 29.6)
In [18]: smap.set_text(91.2, 29.7, 'Lhasa', fontsize=17)
In [19]: smap.visualize()
Out[19]:
{'imshow': <matplotlib.image.AxesImage at 0x7f97fec2eec0>,
'contour': [],
'contourf': []}
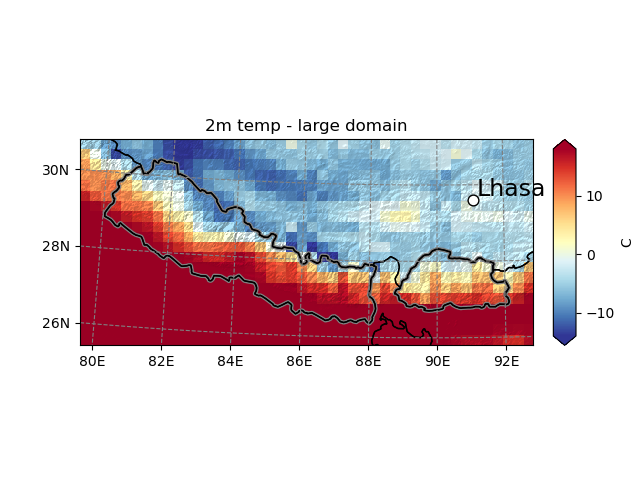
Maps are persistent, which is useful when you have many plots to do. Plotting further data on them is possible, as long as the geolocalisation information is shipped with the data (in that case, the DataArray’s attributes are lost in the conversion from Kelvins to degrees Celsius so we have to set it explicitly):
In [20]: smap.set_data(ds.T2.isel(Time=1)-273.15, crs=ds.salem.grid)
In [21]: smap.visualize(title='2m temp - large domain', cbar_title='C')
Out[21]:
{'imshow': <matplotlib.image.AxesImage at 0x7f97fec2dab0>,
'contour': [],
'contourf': []}
Reprojecting data¶
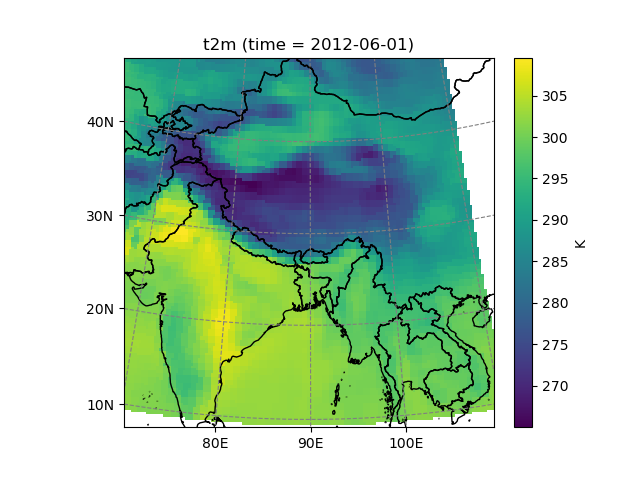
Salem can also transform data from one grid to another:
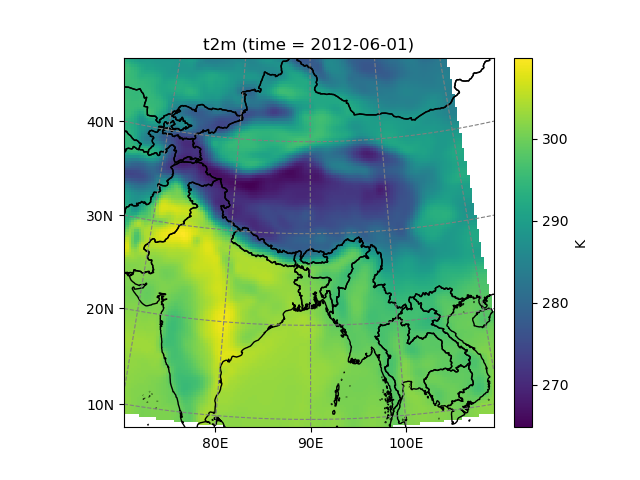
In [22]: dse = salem.open_xr_dataset(get_demo_file('era_interim_tibet.nc'))
In [23]: t2_era_reproj = ds.salem.transform(dse.t2m)
In [24]: assert t2_era_reproj.salem.grid == ds.salem.grid
In [25]: t2_era_reproj.isel(time=0).salem.quick_map()
Out[25]: <salem.graphics.Map at 0x7f97fed1a680>
In [26]: t2_era_reproj = ds.salem.transform(dse.t2m, interp='spline')
In [27]: t2_era_reproj.isel(time=0).salem.quick_map()
Out[27]: <salem.graphics.Map at 0x7f97fed2f2e0>